Unsupervised learning education teaches a different kind of intelligence: not the ability to answer questions, but the ability to notice what questions might exist. When labels are absent, the student cannot lean on “correct answers.” Instead, they learn to search for structure—clusters, trends, latent dimensions—while staying humble about what patterns truly mean. This diagram captures that discipline. The inputs—unlabeled data and feature sets—bring in raw, often messy reality: signals without explanations. The controls—curriculum goals and evaluation criteria—prevent pattern-hunting from turning into story-telling; they require learners to justify choices, test stability, compare alternatives, and avoid mistaking noise for discovery. Inside the central function, students practice the craft of exploration: selecting representations, tuning algorithms, validating clusters, interpreting embeddings, and learning when “interesting structure” is real insight and when it is an artifact of preprocessing. The mechanisms—algorithms and compute resources—provide the practical engine for repeated trials, sensitivity checks, and meaningful comparisons at scale. The outputs are therefore twofold: practitioners who can explore data with care, and discovered structure—like clusters or compact representations—that can later guide decisions, generate hypotheses, or prepare the ground for more targeted modeling.
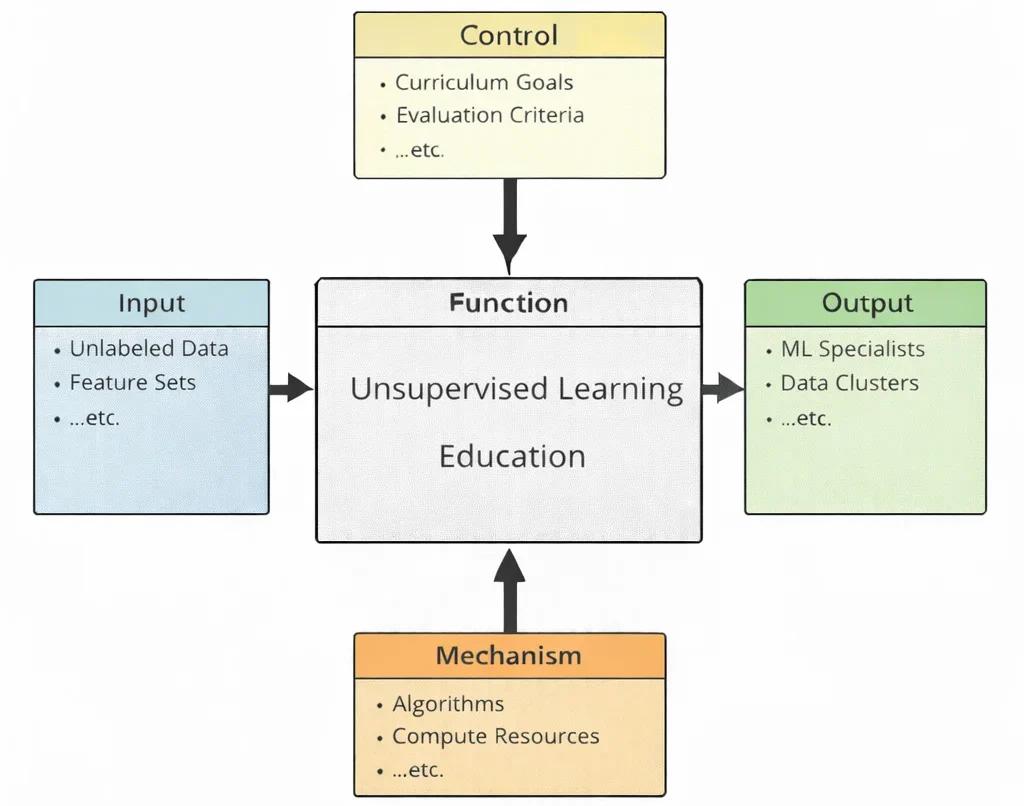
This IDEF0 (Input–Control–Output–Mechanism) diagram summarizes Unsupervised Learning Education as a structured learning process. Inputs on the left—unlabeled data, feature sets, …etc.—represent the core starting point of unsupervised learning: data without provided answers, where the task is to uncover hidden patterns and structure. Controls at the top—curriculum goals, evaluation criteria, …etc.—define what students should learn and how their work is judged, including expectations about validity, interpretability, and methodological rigor. The central function, Unsupervised Learning Education, transforms these inputs and controls into practical understanding through guided study and iterative experimentation. Outputs on the right—ML specialists, data clusters, …etc.—represent intended outcomes: learners who can apply unsupervised methods responsibly, and meaningful groupings or representations that reveal structure in complex data. Mechanisms at the bottom—algorithms, compute resources, …etc.—provide the enabling tools that make discovery possible, from clustering and dimensionality reduction methods to the computational power needed to test and refine results.
Unsupervised learning is a core branch of artificial intelligence and machine learning that finds structure in unlabeled data. Unlike supervised learning, it discovers patterns without predefined outputs—grouping similar items (clustering), compressing signals (dimensionality reduction), and surfacing rare events (anomaly detection). These capabilities are widely used across information technology for segmentation, monitoring, search, and exploration.
In computer vision, unsupervised methods help with image compression, feature learning, and object discovery. In natural language processing (NLP), they power topic modeling and word/sentence embeddings without labeled corpora. Coupled with deep learning, unsupervised and self-supervised pretraining improve models when labeled data is scarce.
Real-world impact spans data science & analytics (exploratory data analysis at scale) and cloud computing (distributed training over large datasets), with flexible cloud deployment models that support iterative clustering and embedding pipelines. These advances align with broader emerging technologies initiatives.
High-value domains like the Internet of Things (IoT) and smart technologies use unsupervised tools to analyze sensor streams and detect usage patterns. In smart manufacturing and Industry 4.0, they help optimize workflows and flag equipment anomalies. Even in satellite technology, unsupervised models classify remote-sensing imagery and uncover environmental change.
At the frontier, unsupervised learning may intersect with quantum computing—leveraging qubits, superposition, and quantum gates for new ways to encode and analyze information—and with space exploration technologies for autonomous classification and navigation.
Connections to reinforcement learning are growing as agents learn behaviors with minimal supervision, while expert systems can mine rules/cases for adaptive updates. These ideas are central to robotics and autonomous systems, which must interpret complex environments and adapt without explicit guidance.
Across digital infrastructure—internet & web technologies, search, and content organization—and the wider STEM landscape, unsupervised learning scales insight as data grows, turning raw signals into structure, profiles, and actions.

This image visualizes unsupervised learning as an exploration of patterns rather than a march toward predefined answers. The luminous, network-like brain represents an algorithm extracting relationships from raw data, while the surrounding charts, color wheels, and grouped points hint at clustering, dimensionality reduction, and feature discovery. Instead of “correct labels,” the scene emphasizes organization: separating data into natural groups, revealing trends, and compressing complex information into simpler representations. The energetic, multi-colored design conveys the idea of models “making sense” of messy datasets by detecting structure, similarity, and emerging categories on their own.
What is unsupervised learning, and how is it different from supervised learning?
Unsupervised learning is a type of machine learning where algorithms are trained on data without labeled outputs, and the goal is to discover hidden patterns, groupings, or structures in the data. In contrast, supervised learning uses labeled examples with known targets to learn a direct mapping from inputs to outputs. Unsupervised learning is especially useful when labels are expensive or unavailable, and when we want to explore data, detect clusters, or reduce dimensionality before building predictive models.
What are the main types of unsupervised learning algorithms?
The main types of unsupervised learning algorithms include clustering methods, dimensionality reduction, association rule learning, and density estimation. Clustering methods such as k-means and hierarchical clustering group similar data points together, dimensionality reduction techniques like principal component analysis (PCA) compress data into fewer informative features, and association rule learning uncovers relationships between variables in large datasets. These algorithms help simplify complex data and reveal structures that are not obvious from raw observations alone.
What are some real-world applications of unsupervised learning?
Unsupervised learning is widely used in customer segmentation, anomaly detection, recommendation systems, and exploratory data analysis. For example, businesses use clustering to group customers with similar behaviors for targeted marketing, and anomaly detection algorithms to flag unusual transactions that may indicate fraud. In recommendation systems and content platforms, unsupervised techniques help discover latent interests and organize large collections of items, making it easier to personalize user experiences.
What is clustering in unsupervised learning, and how do methods like k-means work?
Clustering is an unsupervised learning task where the goal is to group data points so that items in the same cluster are more similar to each other than to those in other clusters. In k-means clustering, we choose a number of clusters, k, and the algorithm iteratively assigns data points to the nearest cluster center and then updates the centers as the mean of their assigned points. This process continues until the assignments stabilize, producing compact clusters that summarize the main groupings in the data.
What is dimensionality reduction, and why is it important in unsupervised learning?
Dimensionality reduction refers to techniques that transform high-dimensional data into a lower-dimensional representation while preserving as much relevant information as possible. Methods such as principal component analysis (PCA), t-SNE, and autoencoders compress data into a smaller set of features that capture key patterns and variability. This is important because it helps reduce noise, visualise complex datasets, speed up learning algorithms, and mitigate the curse of dimensionality, which can make high-dimensional data difficult to analyze directly.
How can we evaluate the quality of an unsupervised learning model without labels?
Evaluating unsupervised learning is challenging because there are no ground-truth labels, but several internal and indirect measures are commonly used. For clustering, metrics such as the silhouette score, within-cluster sum of squares, and cluster separation give insight into how compact and well-separated the groups are. For dimensionality reduction and representation learning, reconstruction error, visualization quality, and downstream performance on supervised tasks can be used to judge whether the learned structure is meaningful and useful.
How does unsupervised learning support other AI and machine learning pipelines?
Unsupervised learning often acts as a foundational step in larger AI pipelines by discovering structure that can be reused in supervised tasks. It can be used for feature extraction, where patterns learned from unlabeled data provide richer inputs for later classification or regression models, and for pretraining deep networks before fine-tuning on labeled data. By organizing data, reducing dimensionality, and revealing key groups, unsupervised learning makes downstream models more efficient, robust, and informative.
What background knowledge is helpful for studying unsupervised learning at university level?
To study unsupervised learning at university level, it is helpful to have a grounding in linear algebra, probability, basic statistics, and introductory machine learning concepts. Familiarity with vector spaces, matrices, eigenvalues, and distributions makes it easier to understand algorithms like PCA and clustering. Programming experience, especially in Python and libraries such as NumPy, scikit-learn, or PyTorch, is also important for implementing algorithms and experimenting with real datasets.
Table of Contents
Core Concepts of Unsupervised Learning
Absence of Labels:
Unlike supervised learning, where each data instance is paired with a known target, unsupervised learning has no such “answer key.” The model must rely entirely on the input features to detect commonalities, differences, and relationships. This is especially useful when labels are expensive or time-consuming to obtain, or when the data’s structure is not well understood.
Discovery of Hidden Structures:
Without explicit guidance, unsupervised models often reveal underlying patterns. These patterns might correspond to meaningful groupings, latent factors, or informative lower-dimensional representations of complex data. This can help researchers, analysts, and organizations understand their data better, leading to more informed decision-making or subsequent application in supervised tasks.
Adaptability and Exploration:
Since there are no labels dictating what “correct” behavior looks like, unsupervised techniques can be highly exploratory. Data scientists frequently use these methods as a first step to characterize and understand a dataset before applying more complex predictive models.
Common Techniques in Unsupervised Learning
Clustering:
Clustering algorithms partition the data into groups (clusters) so that instances within the same cluster are more similar to each other than to those in different clusters.
Customer Segmentation:
For businesses looking to tailor marketing strategies, clustering can identify groups of customers with similar purchasing habits, interests, or demographics. By revealing segments within a customer base, companies can develop targeted campaigns, personalized product recommendations, and optimized service offerings.
Document Grouping:
In text analysis, clustering can organize large collections of documents by topic or style without any prior classification. This helps with tasks like news article categorization, social media content analysis, or internal document management, enabling users to quickly discover patterns in unstructured text.
Common clustering algorithms include k-means, hierarchical clustering, and DBSCAN. Each algorithm has its strengths—k-means is simple and fast, hierarchical clustering reveals nested groupings, and DBSCAN finds clusters of arbitrary shape and can identify outliers.
Dimensionality Reduction:
Real-world datasets often contain a large number of features—some of which may be redundant, noisy, or less informative. Dimensionality reduction techniques help condense this complexity into a more manageable and interpretable form.
Principal Component Analysis (PCA):
PCA identifies the directions (principal components) along which the data varies most. By projecting data onto these components, PCA can reduce the number of features while retaining the bulk of the important information. This simplifies visualization, speeds up computations, and can improve the performance of downstream tasks like clustering or classification.
Feature Extraction and Visualization:
Beyond PCA, other techniques like t-SNE (t-distributed Stochastic Neighbor Embedding) or UMAP (Uniform Manifold Approximation and Projection) help visualize high-dimensional data by placing similar points close together in a two- or three-dimensional space. This can reveal natural groupings, outliers, or complex relationships that might be otherwise hidden.
Evaluation & Validation for Unsupervised Learning
Without labels, “good” structure must be justified with internal indices (use only the data and cluster assignments), stability checks (is the solution reproducible?), and—when you do have labels for auditing or a downstream task—external indices. This section is a practical playbook you can run end-to-end.
1) Internal Indices (no labels required)
- Silhouette score (−1 … 1): cohesion vs separation. Higher is better. Useful to scan
k. - Davies–Bouldin (≥0): average similarity of each cluster with its most similar one. Lower is better.
- Calinski–Harabasz (≥0): between-cluster dispersion vs within-cluster dispersion. Higher is better.
2) External Indices (when reference labels exist)
- Adjusted Rand Index (ARI): agreement of assignments up to permutation; chance-corrected (−1 … 1).
- Normalized Mutual Information (NMI): mutual information normalized to [0, 1]; robust to label permutations.
Use these to sanity-check on synthetic data, curated benchmarks, or when clusters are compared to known categories.
3) Stability & Robustness
- Resampling: bootstraps or 80/20 subsamples; compare assignments via ARI/NMI across runs.
- Perturbations: add small noise, shuffle feature order, or drop a few features; watch metric drift.
- Initialization sensitivity: run multiple random seeds and aggregate scores (mean ± std).
- Silhouette sweep: compute silhouette for k=2…K and pick the local maximum that’s stable across seeds.
- Elbow method: look for diminishing returns in within-cluster SSE vs k.
- Gap statistic: compare dispersion to a null reference distribution (uniform over bounding box).
- Clusterability test: Hopkins statistic > 0.5 suggests non-random structure worth clustering.
4) Quick Python Recipes (scikit-learn)
# pip install scikit-learn numpy matplotlib
import numpy as np
from sklearn.cluster import KMeans
from sklearn.metrics import silhouette_score, davies_bouldin_score, calinski_harabasz_score
from sklearn.metrics import adjusted_rand_score, normalized_mutual_info_score
def evaluate_internal(X, labels):
return {
"silhouette": silhouette_score(X, labels),
"davies_bouldin": davies_bouldin_score(X, labels),
"calinski_harabasz": calinski_harabasz_score(X, labels),
}
def choose_k_by_silhouette(X, k_min=2, k_max=12, n_init=10, seed=42):
rng = np.random.RandomState(seed)
results = []
for k in range(k_min, k_max + 1):
km = KMeans(n_clusters=k, n_init=n_init, random_state=rng).fit(X)
s = silhouette_score(X, km.labels_)
results.append((k, s))
# pick the best k by silhouette (you may also smooth and check stability)
best_k, best_s = max(results, key=lambda t: t[1])
return best_k, results
# Example usage
# X: (n_samples, n_features) numpy array
# y_true: optional labels for external indices
# best_k, sweep = choose_k_by_silhouette(X)
# km = KMeans(n_clusters=best_k, n_init=20, random_state=0).fit(X)
# internal = evaluate_internal(X, km.labels_)
# if y_true is not None:
# external = {
# "ARI": adjusted_rand_score(y_true, km.labels_),
# "NMI": normalized_mutual_info_score(y_true, km.labels_)
# }5) Reporting Best Practices
- Show a table with internal metrics (silhouette, CH, DB) for the final model and ± variation across seeds.
- If labels exist: add external metrics (ARI, NMI) for auditing—clearly labeled “reference-only”.
- Include cluster profiles (feature means/medians, top tokens) and representative exemplars per cluster.
- Discuss stability (resampling/perturbation outcomes) and limit cases (outliers, density skew, scale).
6) Pitfalls to Avoid
- Feature scale mismatch: standardize/whiten where appropriate; use cosine for sparse/high-dim data.
- “Pretty plots ≠ good clusters”: 2-D projections (PCA/UMAP/t-SNE) can distort distances—always pair with metrics.
- Over-interpreting k: multiple k may be “valid”; prefer the simplest story that is stable and useful.
- Leakage into downstream tasks: if these clusters feed a supervised model, fit transforms inside CV folds.
What you get: a defensible selection of k, quantifiable quality, and evidence of robustness—essential for reviews, stakeholders, and production readiness.
Leakage-Safe Pipelines for Unsupervised → Supervised Workflows
Many real projects use unsupervised transforms (e.g., scaling, PCA, clustering, embeddings) to create features, then train a
supervised model (e.g., logistic regression, tree, or boosting). All transforms that learn from data must be
fit only on each fold’s training split—never on the whole dataset—otherwise information leaks from validation into training.
The clean way to guarantee this is to put every learnable step inside a Pipeline and evaluate inside cross-validation.
Typical leakage traps
- Fitting scalers/encoders/PCA on the full dataset, then cross-validating the classifier (leaks validation statistics).
- Running clustering once on all data and passing cluster IDs/distances to the model (leaks cluster structure).
- Pre-computing text/vector embeddings on all data (including validation) and then training a classifier on those vectors.
- For time series, using random CV instead of TimeSeriesSplit (look-ahead leakage).
.fit(...) anywhere, it belongs inside the pipeline;
cross-validation must refit that step from scratch on each fold’s train split.1) Pattern A — Unsupervised features → classifier (scikit-learn)
Standardize + One-Hot → (PCA features ⊕ K-Means distances) → LogisticRegression.
K-Means is valid as a transformer because transform returns distances to cluster centers.
# pip install scikit-learn numpy
import numpy as np
from sklearn.compose import ColumnTransformer
from sklearn.pipeline import Pipeline, FeatureUnion
from sklearn.preprocessing import StandardScaler, OneHotEncoder
from sklearn.decomposition import PCA
from sklearn.cluster import KMeans
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import StratifiedKFold, cross_val_score
# X: DataFrame or ndarray; y: labels
num = ["age","income","tenure"] # numeric columns
cat = ["city","segment"] # categorical columns
pre = ColumnTransformer([
("num", StandardScaler(), num),
("cat", OneHotEncoder(handle_unknown="ignore"), cat)
])
unsup = FeatureUnion([
("pca", PCA(n_components=12, random_state=0)),
("kmeans", KMeans(n_clusters=8, random_state=0)) # transform -> distances (n_clusters features)
])
pipe = Pipeline([
("pre", pre), # refit per fold
("unsup", unsup), # refit per fold (PCA + KMeans)
("clf", LogisticRegression(max_iter=2000))
])
cv = StratifiedKFold(n_splits=5, shuffle=True, random_state=42)
scores = cross_val_score(pipe, X, y, cv=cv, scoring="roc_auc")
print("AUC mean ± std:", scores.mean(), scores.std())2) Pattern B — Tune unsupervised + supervised pieces together
Grid search the whole pipeline. Parameters are referenced by step names, e.g. unsup__pca__n_components.
from sklearn.model_selection import GridSearchCV
param_grid = {
"unsup__pca__n_components": [8, 12, 20],
"unsup__kmeans__n_clusters": [6, 8, 12],
"clf__C": [0.1, 1.0, 3.0]
}
search = GridSearchCV(pipe, param_grid=param_grid, cv=cv, scoring="roc_auc", n_jobs=-1)
search.fit(X, y)
print("Best AUC:", search.best_score_)
print("Best params:", search.best_params_)3) Pattern C — Time-series (no look-ahead)
Use TimeSeriesSplit so that each validation fold strictly follows its training window in time.
from sklearn.model_selection import TimeSeriesSplit
tscv = TimeSeriesSplit(n_splits=5)
scores = cross_val_score(pipe, X, y, cv=tscv, scoring="neg_log_loss")
print("TS log-loss mean ± std:", scores.mean(), scores.std())4) Using external embeddings (text/images)
- If the embedding model is frozen and trained on a separate large corpus (e.g., a public model), it’s usually fine to use it as a fixed deterministic transform. Still, compute embeddings only from the fold’s training data where possible.
- If you train or fine-tune the embedding model on your dataset, it must be done inside each fold (or with nested CV) to avoid peeking.
- Print the CV object you used (
StratifiedKFold/TimeSeriesSplit) and confirm the pipeline contains all learnable steps (scalers, encoders, PCA, KMeans, vectorizers). - Rerun with different
random_statevalues; variance should be small and conclusions unchanged. - Plot feature importances or coefficients of the final model to confirm they’re stable across folds.
- For time series, audit a few fold splits by timestamps to ensure no future data leaks backward.
5) Minimal template you can copy/paste
# Template: place every learnable step inside the Pipeline
from sklearn.compose import ColumnTransformer
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler, OneHotEncoder
from sklearn.decomposition import PCA
from sklearn.cluster import KMeans
from sklearn.linear_model import LogisticRegression
pre = ColumnTransformer([
("num", StandardScaler(), num_cols),
("cat", OneHotEncoder(handle_unknown="ignore"), cat_cols)
])
pipe = Pipeline([
("pre", pre),
("pca", PCA(n_components=12, random_state=0)),
("kmeans", KMeans(n_clusters=8, random_state=0)),
("clf", LogisticRegression(max_iter=2000))
])
# Evaluate with cross_val_score/GridSearchCV; for time series, use TimeSeriesSplit.Takeaway: Pipelines + CV treat unsupervised transforms as first-class citizens. That keeps your metrics honest,
your model reproducible, and your production path simple (save one fitted pipeline; reload and call .predict).
Interpreting Clusters: Profiles, Exemplars & Drift Monitoring
Clustering only becomes valuable once teams can explain segments, use them downstream, and monitor them over time. This block shows how to (1) build human-readable profiles, (2) select representative exemplars, (3) validate separation, and (4) watch for drift in production.
1) Build human-readable cluster profiles
After assigning cluster labels on the training split, aggregate key features to describe each segment (means, medians, rates).
# X_tr: training features (DataFrame), labels_tr: KMeans().fit_predict(X_tr)
import pandas as pd
def cluster_profile_table(X_tr: pd.DataFrame, labels_tr, top_n_cat=5):
df = X_tr.copy()
df["cluster"] = labels_tr
prof = df.groupby("cluster").agg(["mean","median","std"])
# Optionally add categorical prevalences
cat_cols = [c for c in df.columns if df[c].dtype == "object"]
for c in cat_cols[:top_n_cat]:
prev = (df.groupby("cluster")[c]
.value_counts(normalize=True)
.rename("prev")
.reset_index())
# keep the top category per cluster
prev = prev.loc[prev.groupby("cluster")["prev"].idxmax()].set_index("cluster")
prof[(c,"top_cat")] = prev.set_index(prev.index)["level_1"]
prof[(c,"top_prev")] = prev.set_index(prev.index)["prev"]
return prof
profiles = cluster_profile_table(X_train_df, kmeans_labels_train)
print(profiles.head())2) Pick exemplars (most representative points)
Select the closest data points to each centroid—great for design reviews, annotation, or UI previews.
import numpy as np
from sklearn.metrics import pairwise_distances_argmin_min
# kmeans is the fitted model on training data
assign = kmeans.labels_
closest_idx, dists = pairwise_distances_argmin_min(X_train_df.values, kmeans.cluster_centers_)
# closest_idx gives the index of the nearest centroid for each row; we want top-k per cluster:
def topk_exemplars(X, labels, centers, k=3):
out = {}
for c in range(centers.shape[0]):
inds = np.where(labels==c)[0]
_, d = pairwise_distances_argmin_min(X.values[inds], centers[c].reshape(1,-1))
top = inds[np.argsort(d)[:k]]
out[c] = top
return out
exemplars = topk_exemplars(X_train_df, assign, kmeans.cluster_centers_, k=3)3) Explain why points fall into clusters
A practical trick is to train a simple surrogate classifier to predict the cluster label and inspect its coefficients/feature importances.
# Surrogate: multinomial logistic regression over cluster IDs
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import classification_report
from sklearn.model_selection import StratifiedKFold, cross_val_score
y_sur = assign # cluster id as target
sur = LogisticRegression(max_iter=2000, multi_class="multinomial")
cv = StratifiedKFold(n_splits=5, shuffle=True, random_state=42)
print("Surrogate accuracy:", cross_val_score(sur, X_train_df, y_sur, cv=cv).mean())
sur.fit(X_train_df, y_sur)
coefs = pd.DataFrame(sur.coef_, columns=X_train_df.columns)
# For each cluster row, the top +/- coefficients indicate features pushing membership.4) Validate separation (are clusters “real”)?
- Silhouette score > 0.30 often signals useful structure (context matters).
- Elbow/inertia flattening suggests diminishing returns in k.
- Check stability: re-run clustering with different seeds, compare label agreement (ARI/NMI).
from sklearn.metrics import silhouette_score
from sklearn.metrics.cluster import adjusted_rand_score
sil = silhouette_score(X_train_df, assign)
print("Silhouette:", round(sil, 3))
# Stability check (quick demo)
assign2 = KMeans(n_clusters=kmeans.n_clusters, n_init=10, random_state=7).fit_predict(X_train_df)
print("ARI vs. new run:", adjusted_rand_score(assign, assign2))5) Monitor drift in production
Track whether live data still “looks like” training data and whether cluster structure shifts.
- Centroid drift: L2 distance between live centroids and training centroids.
- Population mix: % of points per cluster—watch for sudden shifts.
- Distribution drift: PSI/JS distance for key features vs. training baselines.
# 5a) Centroid drift
import numpy as np
def centroid_drift(train_centers, live_centers):
return np.linalg.norm(train_centers - live_centers, axis=1) # per-cluster L2
# 5b) Jensen–Shannon distance between two discrete distributions
import numpy as np
from scipy.spatial.distance import jensenshannon
def js_distance(p, q, eps=1e-12):
p = np.clip(p, eps, 1); p = p/p.sum()
q = np.clip(q, eps, 1); q = q/q.sum()
return jensenshannon(p, q) # 0..1
# 5c) PSI (Population Stability Index) for numeric feature
import numpy as np
def psi(expected, actual, bins=10):
# bin on expected percentiles
cuts = np.percentile(expected, np.linspace(0,100,bins+1))
e, _ = np.histogram(expected, bins=cuts)
a, _ = np.histogram(actual, bins=cuts)
e = np.where(e==0, 1, e); a = np.where(a==0, 1, a)
pe, pa = e/e.sum(), a/a.sum()
return np.sum((pa-pe)*np.log(pa/pe))
# Example thresholds (tune for your domain):
# JS distance > 0.15 or PSI > 0.2 or any centroid drift in top 5% historic -> alert- Log profiles, centroids, and per-cluster counts to your experiment tracker (e.g., MLflow).
- Alert on spikes in centroid drift / PSI / JS distance or abrupt cluster size changes.
- Re-fit clusters on a rolling window if drift persists; keep a champion/challenger setup.
- Version your pipelines and the feature schema; block deployments on schema mismatches.
Takeaway: Profiles make segments understandable, exemplars make them tangible, and drift metrics keep them honest in production.
Dimensionality Reduction Cheat-Sheet (PCA · UMAP · t-SNE)
Use dimensionality reduction to compress features, denoise data, and visualize structure. Choose the tool that matches your goal, data type, and need for speed vs. fidelity.
PCA (Principal Component Analysis)
Linear, orthogonal directions of max variance.
- Pros: fast, deterministic, invertible; useful for compression & denoising.
- Cons: linear only; can miss nonlinear structure/manifolds.
- Good for: feature compression before clustering/regression; anomaly baselines.
- Key knobs:
n_components(variance target or fixed), scale features first.
UMAP (Uniform Manifold Approximation and Projection)
Graph-based, preserves local & some global structure; fast embeddings.
- Pros: often clearer clusters than t-SNE; faster; works well for 2D/3D viz and as features.
- Cons: stochastic; results depend on seeds & parameters.
- Good for: visual exploration, pre-clustering, low-dim features for downstream tasks.
- Key knobs:
n_neighbors(local vs. global),min_dist(cluster tightness),metric.
t-SNE (t-Stochastic Neighbor Embedding)
Nonlinear viz that excels at local neighborhood separation.
- Pros: strong cluster separation in 2D/3D; great for presentations.
- Cons: slow on large data; poor global distances; not ideal as model features.
- Good for: exploratory plots & sanity checks; not for production features.
- Key knobs:
perplexity(~5–50),learning_rate,init,n_iter.
transform validation/test.
If you use CV, refit the reducer inside each fold (e.g., scikit-learn Pipeline).Quick, leakage-safe examples
# PCA (scikit-learn) — compression for clustering/regression
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
pca_pipe = Pipeline([
("scaler", StandardScaler()), # fit on train only
("pca", PCA(n_components=0.95)) # keep 95% variance
])
X_tr_pca = pca_pipe.fit_transform(X_train) # fit here
X_te_pca = pca_pipe.transform(X_test) # transform only# UMAP (umap-learn) — low-dim features for clustering/viz
# pip install umap-learn
import umap
umap_pipe = Pipeline([
("scaler", StandardScaler()),
("umap", umap.UMAP(n_neighbors=15, min_dist=0.1, n_components=2, random_state=42))
])
X_tr_umap = umap_pipe.fit_transform(X_train)
X_te_umap = umap_pipe.transform(X_test)# t-SNE (viz only; not ideal as features)
from sklearn.manifold import TSNE
tsne = TSNE(n_components=2, perplexity=30, learning_rate="auto", init="pca",
n_iter=1000, random_state=42, verbose=1)
X_tr_tsne = tsne.fit_transform(StandardScaler().fit_transform(X_train))Which should I use?
- Need speed, compression, interpretability? Start with PCA (variance explained & loadings).
- Need clearer clusters & usable low-dim features? Try UMAP (then cluster on UMAP-2D/10D).
- Need a “wow” plot for local structure? Use t-SNE for visualization only.
Pitfalls & sanity checks
- Scaling: always standardize (or use appropriate metric) before PCA/UMAP/t-SNE.
- Stochasticity: UMAP/t-SNE vary with seed; fix
random_statefor reproducibility. - Global distances: t-SNE distances are not global; don’t infer cluster-to-cluster proximity.
- Over-interpretation: low-dim plots are maps, not ground truth. Cross-check with metrics.
- Large N: downsample for t-SNE; UMAP handles larger N but still watch runtime.
Evaluating embeddings
- Trustworthiness/continuity: how well neighborhoods are preserved in 2D/3D.
- Clustering quality on embeddings: silhouette/ARI/NMI after K-Means/DBSCAN on PCA/UMAP space.
- Downstream lift: train a simple classifier/regressor on PCA/UMAP features; compare baseline.
Supervised follow-ups (turn embeddings into value)
- Clustering → actions: build profiles on PCA/UMAP space, then target with rules/experiments.
- Anomaly detection: IsolationForest/LOF on PCA components to reduce noise & false alarms.
- Classification: feed PCA/UMAP features to linear/logistic models for fast, robust baselines.
Takeaway: Start with PCA for compression & baselines, use UMAP for structure and features, and keep t-SNE for exploration/communication. Always fit reducers on train only and track drift over time.
Anomaly Detection in Practice (Isolation Forest · LOF · One-Class SVM)
Anomaly detection flags rare, unusual, or risky patterns without labels. It’s pivotal in fraud, manufacturing QA, ops monitoring, and security. This section covers when to use which method, how to set thresholds, and how to validate with or without labels.
Method chooser
- Isolation Forest — robust default for tabular; scales well; set
contaminationto target alert rate. - Local Outlier Factor (LOF) — density-based; good for local anomalies; sensitive to
n_neighbors. - One-Class SVM — flexible boundary; powerful but sensitive to kernel/ν; slower on large N.
- Robust Covariance (EllipticEnvelope) — works if data is roughly Gaussian/elliptical.
Preprocessing rules
- Standardize numeric features; one-hot encode categoricals (or use target-free encoders).
- Optionally reduce with PCA (10–50D) to denoise before the detector (fit on train only).
- Remove obvious data glitches upstream; anomalies ≠ bad data ingest.
Leakage-safe pipelines (scikit-learn)
# pip install scikit-learn numpy
import numpy as np
from sklearn.compose import ColumnTransformer
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler, OneHotEncoder
from sklearn.decomposition import PCA
from sklearn.ensemble import IsolationForest
from sklearn.neighbors import LocalOutlierFactor
numeric = ["num_1","num_2", "..."]
categorical = ["cat_1","cat_2", "..."]
pre = ColumnTransformer([
("num", StandardScaler(), numeric),
("cat", OneHotEncoder(handle_unknown="ignore"), categorical)
])
# Pattern A: PCA -> Isolation Forest (predict on new data)
iso_pipe = Pipeline([
("pre", pre),
("pca", PCA(n_components=20, random_state=42)),
("iso", IsolationForest(n_estimators=300, contamination=0.01, random_state=42))
])
# Fit on TRAIN only; evaluate on VAL/TEST
iso_pipe.fit(X_train)
# Scores: higher = more normal in sklearn; get anomaly score as negative score_samples
scores_val = -iso_pipe.score_samples(X_val) # larger = more anomalous
thr = np.percentile(scores_val, 99) # ~1% alert rate; tune
alerts = scores_val >= thr
# Pattern B: LOF as a detector — use novelty=True to allow predict on new data
lof_pipe = Pipeline([
("pre", pre),
("pca", PCA(n_components=20, random_state=42)),
("lof", LocalOutlierFactor(n_neighbors=35, novelty=True))
])
lof_pipe.fit(X_train) # fit on TRAIN
lof_scores = -lof_pipe.score_samples(X_val) # larger = more anomalous
If you have some labels (or can label a sample)
- Use PR-AUC (imbalanced) and AUROC on a temporally separated validation set.
- Report precision@k (k = daily review capacity) and cost curves (false-alarm burden).
- Keep a small human-in-the-loop review queue; feed adjudicated results back as weak labels.
# Example metrics when labels exist (y=1 anomaly, y=0 normal)
from sklearn.metrics import average_precision_score, roc_auc_score, precision_recall_curve
y_val = y_val_labels
ap = average_precision_score(y_val, scores_val)
roc = roc_auc_score(y_val, scores_val)
print("PR-AUC:", round(ap,3), "AUROC:", round(roc,3))
# Precision@K (top K scored examples)
K = 100
topk = np.argsort(scores_val)[-K:]
prec_at_k = y_val[topk].mean()
print("Precision@{}:".format(K), round(prec_at_k,3))
No labels? Three practical validations
- Stability: re-fit with new seeds; Spearman correlation between score vectors should be high.
- Perturbations: small noise/drop-feature tests; anomaly rankings shouldn’t flip wildly.
- Spot-check: review top-N alerts with SMEs; log acceptance rate and adjust thresholds.
When to prefer other tools
- Sequences / time series: use sliding windows with feature stats; consider HMMs or autoencoders.
- Images: use reconstruction-error from an autoencoder or VAE as the anomaly score (fit on normals).
- Graphs: consider graph LOF / subgraph frequency; watch scalability.
Takeaway: choose a detector that fits your data, calibrate to operations, validate with stability and (if possible) labels, then monitor alert quality over time.
Self-Supervised Learning (SSL) for Stronger Unsupervised Features
Self-supervised learning creates labels from the data itself, letting you pretrain encoders on large, unlabeled corpora. You then reuse the embeddings for clustering, anomaly detection, or lightweight classifiers (“linear probes”). SSL typically improves separation, stability, and downstream accuracy—especially when labeled data is scarce.
Two core families
- Contrastive learning (e.g., SimCLR/MoCo): push two augmented views of the same item together and push different items apart. Requires strong data augmentations.
- Masked/denoising modeling (e.g., BERT-style MLM, MAE for images): hide parts of input and train the model to reconstruct or predict them.
When to use SSL
- Few or no labels; large unlabeled pool.
- Unclear clusters with raw features; need better representation.
- Domain shift vs public embeddings; pretrain on your own data to capture local structure.
Leakage-safe practice
- Pretrain SSL on train split only (or on an external public corpus). Never let validation/test flow into pretraining.
- Freeze or lightly fine-tune the encoder inside a Pipeline; evaluate in cross-validation or with a proper time split.
Quick recipes
Minimal patterns you can adapt; keep SSL pretraining separate from evaluation.
A) Contrastive image SSL (PyTorch, SimCLR-style)
# pip install torch torchvision
import torch, torch.nn as nn, torch.nn.functional as F
import torchvision.transforms as T
import torchvision.models as models
# 1) Two random augmentations per image
augment = T.Compose([
T.RandomResizedCrop(224, scale=(0.5,1.0)),
T.RandomHorizontalFlip(),
T.ColorJitter(0.4,0.4,0.4,0.1),
T.RandomGrayscale(p=0.2),
T.ToTensor()
])
# 2) Encoder + projection head
encoder = models.resnet18(weights=None)
encoder.fc = nn.Identity()
proj = nn.Sequential(nn.Linear(512,256), nn.ReLU(inplace=True), nn.Linear(256,128))
tau = 0.2 # temperature
def simclr_step(x_batch):
# x1, x2 are two augmented views (N,C,H,W)
z1 = proj(encoder(x_batch[0])); z2 = proj(encoder(x_batch[1]))
z1 = F.normalize(z1, dim=1); z2 = F.normalize(z2, dim=1)
N = z1.size(0)
reps = torch.cat([z1,z2], dim=0) # (2N, d)
sim = reps @ reps.t() / tau # cosine scaled
mask = torch.eye(2*N, dtype=torch.bool, device=sim.device)
sim = sim.masked_fill(mask, -1e9) # remove self-similarity
# positives: (i <-> i+N) and (i+N <-> i)
pos = torch.cat([torch.arange(N,2*N), torch.arange(0,N)]).to(sim.device)
targets = pos
loss = F.cross_entropy(sim, targets)
return loss
# Train: loop over batches of paired augmentations, optimize encoder+proj.
# Save encoder weights; downstream: freeze encoder, extract embeddings, then cluster.B) Masked language modeling (Transformers)
# pip install transformers datasets
from transformers import AutoTokenizer, AutoModelForMaskedLM, DataCollatorForLanguageModeling, Trainer, TrainingArguments
from datasets import load_dataset
tokenizer = AutoTokenizer.from_pretrained("bert-base-uncased")
model = AutoModelForMaskedLM.from_pretrained("bert-base-uncased")
ds = load_dataset("text", data_files={"train": "corpus_train.txt"}) # your unlabeled text (train only)
def tok(ex): return tokenizer(ex["text"], truncation=True, padding="max_length", max_length=128)
tok_ds = ds.map(tok, batched=True, remove_columns=["text"])
collator = DataCollatorForLanguageModeling(tokenizer=tokenizer, mlm=True, mlm_probability=0.15)
args = TrainingArguments(output_dir="./mlm_out", per_device_train_batch_size=32, num_train_epochs=2)
trainer = Trainer(model=model, args=args, train_dataset=tok_ds["train"], data_collator=collator)
trainer.train()
# Downstream: use the encoder's hidden states as embeddings → cluster/classify.- Linear probe: freeze encoder, train a simple logistic/linear model on top with small labeled subset.
- k-NN on embeddings: quick accuracy/recall proxy; stable = good representations.
- Unsupervised: silhouette/CH/DB indices after K-Means on embeddings vs. raw features.
Good augmentations (crucial for contrastive)
- Images: random crops, flips, color jitter, blur, grayscale; avoid distortions that erase class signal.
- Text: span masking, token dropout, sentence reordering (lightly); preserve semantics.
- Tabular: mild noise, dropout, mixup within class-agnostic ranges; keep units/scales consistent.
Common pitfalls
- Too weak/strong augmentations: weak → trivial task; strong → different semantics.
- Leakage: pretraining on full dataset including validation/test (don’t!).
- Small batch for contrastive: batch size acts like #negatives; use memory banks if needed.
- No downstream check: always verify embeddings improve clustering or anomaly precision/recall.
Takeaway: SSL lets you learn task-agnostic, domain-specific features that make clustering cleaner and anomalies crisper—without labels. Keep pretraining leakage-safe, validate with simple probes, and wire embeddings into your existing pipelines.
Linear-Probe Evaluation (Fast Check for SSL/Embeddings)
A linear probe freezes your encoder and trains a tiny classifier on top of the embeddings. If the embeddings are good, a simple model (logistic / k-NN) should achieve solid validation scores. Use this alongside your unsupervised metrics (silhouette/ARI) for a complete picture.
Inputs (assumptions)
E_tr,E_va: embedding matrices for train/validation (encoder trained on train only).y_tr,y_va: labels for a small labeled subset (optional but recommended for a probe).
A) Logistic-regression probe (5×CV + hold-out)
# pip install scikit-learn numpy
import numpy as np
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import StratifiedKFold, cross_val_score
from sklearn.metrics import accuracy_score, f1_score, classification_report
# E_tr, E_va: np.ndarray; y_tr, y_va: 1D arrays (labels)
probe = Pipeline([
("scale", StandardScaler(with_mean=True)),
("clf", LogisticRegression(max_iter=5000, n_jobs=-1))
])
cv = StratifiedKFold(n_splits=5, shuffle=True, random_state=42)
cv_scores = cross_val_score(probe, E_tr, y_tr, cv=cv, scoring="f1_macro", n_jobs=-1)
print("Linear-probe F1_macro (5xCV):", round(cv_scores.mean(), 4), "±", round(cv_scores.std(), 4))
# Fit on full train embeddings, then evaluate on validation
probe.fit(E_tr, y_tr)
pred = probe.predict(E_va)
print("VALID Acc:", round(accuracy_score(y_va, pred), 4),
"F1_macro:", round(f1_score(y_va, pred, average="macro"), 4))
print(classification_report(y_va, pred))B) k-NN probe (quick sanity)
from sklearn.neighbors import KNeighborsClassifier
knn_probe = Pipeline([
("scale", StandardScaler(with_mean=True)),
("knn", KNeighborsClassifier(n_neighbors=5))
])
knn_probe.fit(E_tr, y_tr)
pred_knn = knn_probe.predict(E_va)
print("kNN VALID Acc:", round(accuracy_score(y_va, pred_knn), 4),
"F1_macro:", round(f1_score(y_va, pred_knn, average="macro"), 4))C) Unsupervised quality on embeddings (silhouette + ARI/NMI if labels exist)
from sklearn.cluster import KMeans
from sklearn.metrics import silhouette_score, adjusted_rand_score, normalized_mutual_info_score
K = len(np.unique(y_tr)) if 'y_tr' in locals() else 6 # choose K sensibly
km = KMeans(n_clusters=K, n_init="auto", random_state=42).fit(E_tr)
labels_val = KMeans(n_clusters=K, init=km.cluster_centers_, n_init=1).fit_predict(E_va)
sil = silhouette_score(E_va, labels_val)
print("Silhouette on embeddings:", round(sil, 3))
# If you have ground-truth labels for validation:
if 'y_va' in locals():
ari = adjusted_rand_score(y_va, labels_val)
nmi = normalized_mutual_info_score(y_va, labels_val)
print("ARI:", round(ari, 3), "NMI:", round(nmi, 3))D) Baseline comparison (ensure the lift is real)
Run the same probes on a simple baseline (e.g., StandardScaler → PCA(0.95) on raw features). Your SSL embeddings should match or exceed those scores; otherwise adjust augmentations, training length, or batch size.
Mini Playbook: Raw Data → Embeddings → Clusters → Business Actions
A practical, leakage-safe path to extract structure from unlabeled data and turn it into decisions, campaigns, or alerts. Works with tabular, text (after vectorization), or image embeddings.
1–2 Data hygiene & leakage-safe pipeline
- Impute missing values by type; cap/transform heavy tails if needed.
- Use ColumnTransformer to scale numeric & encode categorical features.
- Rule: fit all transformers on train only; refit inside each CV fold.
# scikit-learn leakage-safe skeleton
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import StandardScaler, OneHotEncoder
from sklearn.pipeline import Pipeline
numeric = ["num_1","num_2", ...]
categorical = ["cat_1","cat_2", ...]
pre = ColumnTransformer(
transformers=[
("num", StandardScaler(), numeric),
("cat", OneHotEncoder(handle_unknown="ignore"), categorical)
],
remainder="drop"
)
# pre.fit(X_train); X_tr = pre.transform(X_train) ... (prefer inside a Pipeline)3 Embeddings via PCA or UMAP
- PCA: fast/easy for compression; log explained_variance_ratio_.
- UMAP: clearer local structure; log n_neighbors, min_dist, metric.
- Start with 10–20D for clustering, or 2D/3D for visualization.
# Example: PCA → 95% variance
from sklearn.decomposition import PCA
emb = PCA(n_components=0.95, random_state=42)4 Clustering on embeddings
- K-Means: compact, spherical groups; pick K via silhouette or elbow.
- DBSCAN: density-based; finds outliers; choose eps/min_samples.
- GMM: soft assignments; captures ellipses; compare BIC/AIC.
# K-Means after reduction
from sklearn.cluster import KMeans
kmeans = KMeans(n_clusters=6, n_init="auto", random_state=42)5 Validate, profile & monitor
- Quality: silhouette, Davies–Bouldin, stability across seeds/bootstraps.
- Profiles: per-cluster means/medians, top categorical modes, exemplar points.
- Drift: track cluster counts, feature means, PCA variance, UMAP trustworthiness.
# Silhouette on embeddings
from sklearn.metrics import silhouette_score
score = silhouette_score(X_emb_val, labels_val)End-to-end Pipeline (PCA → K-Means), leakage-safe
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import StandardScaler, OneHotEncoder
from sklearn.decomposition import PCA
from sklearn.cluster import KMeans
from sklearn.pipeline import Pipeline
from sklearn.metrics import silhouette_score
import numpy as np
pre = ColumnTransformer([
("num", StandardScaler(), numeric),
("cat", OneHotEncoder(handle_unknown="ignore"), categorical)
])
pipe = Pipeline([
("pre", pre),
("pca", PCA(n_components=0.95, random_state=42)),
("km", KMeans(n_clusters=6, n_init="auto", random_state=42))
])
# Fit only on train, transform val/test:
pipe.fit(X_train)
X_val_emb = pipe.named_steps["pca"].transform(pipe.named_steps["pre"].transform(X_val))
labels_val = pipe.named_steps["km"].predict(pipe.named_steps["pca"].transform(
pipe.named_steps["pre"].transform(X_val)))
sil = silhouette_score(X_val_emb, labels_val)n_components (PCA) or n_neighbors/min_dist (UMAP) and
n_clusters for K-Means; pick by validation silhouette or stability.Action design patterns (turn clusters into value)
- Segmentation → targeting: map clusters to messaging/offers; A/B test lift vs. baseline.
- Anomaly triage: DBSCAN noise points → manual review or automated alerts.
- Product discovery: cluster embeddings of items or sessions → recommendations/menus.
- Ops rules: define simple, human-readable rules from cluster profiles for frontline teams.
Quick chooser
- If you need compression + speed: PCA → K-Means.
- If you want clearer structure/features: UMAP → K-Means (use 2D for viz, 10–20D for clustering).
- If you expect noise/outliers: PCA/UMAP → DBSCAN.
- If clusters overlap/are elongated: PCA → GMM.
Takeaway: keep the pipeline leakage-safe, reduce to a useful embedding, cluster, then validate and convert profiles into concrete actions with A/B tests and monitoring.
Retraining & Monitoring (Cron Sketch)
A lightweight schedule and job outline to keep unsupervised segments healthy in production.
Nightly Metrics & Alerts
- Cluster mix drift (live vs. baseline): PSI >
0.2→ warn; >0.3→ alert. - Centroid drift (per-cluster L2): any in top 5% historic → warn; top 1% → alert.
- Embedding drift (JS distance on PCA comp. or key features): JS >
0.15→ warn; >0.25→ alert. - Silhouette (validation sample): drop >
0.05vs. last week → warn; >0.1→ alert.
Weekly Review & Retrain
- Recompute PCA explained variance (watch for −>
> 5%drop). - Stability check: re-run clustering with new seed; ARI <
0.8→ consider re-fit. - If 2+ alerts fired this week → retrain pipeline (same parameters) and compare metrics A/B.
- Monthly: small grid on
n_components,n_clusters; pick by validation silhouette/stability.
Minimal nightly job (Python skeleton)
# pip install scikit-learn numpy scipy joblib pandas
import numpy as np, pandas as pd, joblib, json, datetime as dt
from sklearn.metrics import silhouette_score
from scipy.spatial.distance import jensenshannon
# --- load fitted pipeline (pre -> pca -> kmeans) and baselines ---
pipe = joblib.load("pipeline_pca_kmeans.joblib")
baseline = json.load(open("baseline_metrics.json")) # stores centroids, cluster_props, pca_var, thresholds
# --- helper metrics ---
def psi(expected, actual, bins=10):
cuts = np.percentile(expected, np.linspace(0,100,bins+1))
e,_ = np.histogram(expected, bins=cuts); a,_ = np.histogram(actual, bins=cuts)
e = np.where(e==0,1,e); a = np.where(a==0,1,a)
pe, pa = e/e.sum(), a/a.sum()
return float(np.sum((pa-pe)*np.log(pa/pe)))
def js_distance(p, q, eps=1e-12):
p = np.clip(p, eps, 1); p = p/p.sum()
q = np.clip(q, eps, 1); q = q/q.sum()
return float(jensenshannon(p, q)) # 0..1
def centroid_drift(train_centers, live_centers):
return np.linalg.norm(train_centers - live_centers, axis=1).tolist()
# --- fetch fresh slice (e.g., last 24h) ---
X_live = pd.read_parquet("live_last24h.parquet") # replace with your source
# --- transform & assign ---
Z_live = pipe.named_steps["pre"].transform(X_live)
E_live = pipe.named_steps["pca"].transform(Z_live)
labels_live = pipe.named_steps["km"].predict(E_live)
# --- compute metrics ---
# 1) cluster mix drift
k = pipe.named_steps["km"].n_clusters
counts = np.bincount(labels_live, minlength=k)
props_live = counts / counts.sum()
props_base = np.array(baseline["cluster_props"])
mix_js = js_distance(props_base, props_live)
# 2) centroid drift
km = pipe.named_steps["km"]
live_centers = []
for c in range(k):
idx = np.where(labels_live==c)[0]
if len(idx)==0:
live_centers.append(km.cluster_centers_[c]) # fallback
else:
live_centers.append(E_live[idx].mean(axis=0))
live_centers = np.vstack(live_centers)
cent_drift = centroid_drift(np.array(baseline["centroids"]), live_centers)
# 3) embedding drift on first few PCA comps
p_base = np.array(baseline["pca_means"]) # per-component mean from training
p_live = E_live.mean(axis=0)
emb_js = js_distance(np.abs(p_base), np.abs(p_live)) # crude summary; customize per feature
# 4) silhouette on a sample (guard for very small N)
sil = float("nan")
if len(X_live) >= 100 and len(np.unique(labels_live)) > 1:
# sample to keep runtime low
idx = np.random.RandomState(0).choice(len(E_live), size=min(5000, len(E_live)), replace=False)
sil = float(silhouette_score(E_live[idx], labels_live[idx]))
# --- thresholds ---
TH = baseline["thresholds"]
alerts = []
if mix_js > TH["mix_js_warn"]: alerts.append({"level":"warn","what":"cluster_mix_js","value":mix_js})
if mix_js > TH["mix_js_alert"]: alerts.append({"level":"alert","what":"cluster_mix_js","value":mix_js})
if max(cent_drift) > TH["centroid_drift_warn"]: alerts.append({"level":"warn","what":"centroid_drift_max","value":max(cent_drift)})
if max(cent_drift) > TH["centroid_drift_alert"]: alerts.append({"level":"alert","what":"centroid_drift_max","value":max(cent_drift)})
if not np.isnan(sil):
if (baseline["silhouette_weekly"] - sil) > TH["silhouette_drop_warn"]:
alerts.append({"level":"warn","what":"silhouette_drop","value":sil})
if (baseline["silhouette_weekly"] - sil) > TH["silhouette_drop_alert"]:
alerts.append({"level":"alert","what":"silhouette_drop","value":sil})
# --- log & (optionally) notify ---
stamp = dt.datetime.utcnow().isoformat()
row = {
"ts": stamp, "mix_js": mix_js, "centroid_drift_max": max(cent_drift),
"emb_js": emb_js, "silhouette": sil, "cluster_props": props_live.tolist(),
"alerts": alerts
}
print(json.dumps(row, indent=2))
pd.DataFrame([row]).to_csv("ul_nightly_log.csv", mode="a", index=False, header=False)
# TODO: send webhook/email if any alert; rotate baselines weekly after manual sign-off.Set up the cron
# Run nightly at 02:15 UTC (example)
15 2 * * * /usr/bin/python3 /path/to/nightly_ul_monitor.py >> /var/log/ul_monitor.log 2>&1
# Run weekly retrain every Sunday at 03:00 UTC (guarded by alerts count)
0 3 * * 0 /usr/bin/python3 /path/to/weekly_ul_retrain.py >> /var/log/ul_retrain.log 2>&1Takeaway: Small, consistent checks (mix, centroids, embeddings, silhouette) + clear thresholds keep unsupervised systems healthy and trustworthy. Retrain only when drift persists or quality drops materially.
Practical Considerations:
Choosing the Number of Clusters or Components:
Unsupervised methods often require assumptions about parameters, such as the number of clusters (k) in k-means or the number of principal components in PCA. Determining these values can be challenging. Methods like the “elbow method” for clustering or examining explained variance for PCA can guide users in making informed choices.
Handling Outliers and Noise:
Unsupervised models can be sensitive to outliers or noisy data. For example, a few extreme values might skew the results of clustering algorithms or obscure the structure revealed by dimensionality reduction. Careful preprocessing—cleaning data, removing obvious errors, or applying robust scaling—helps ensure more reliable outcomes.
Interpreting Results:
ince there are no labels to confirm if the patterns found are “correct,” interpreting the clusters or components requires domain knowledge. Analysts must consider their domain’s context, the meaning of the features, and possible confounding factors. Validating insights with expert opinions, or combining unsupervised findings with external data, can make results more actionable.
Integrations with Other Machine Learning Approaches
Unsupervised learning often serves as a stepping stone:
Preprocessing for Supervised Tasks:
After using unsupervised methods to reduce dimensionality or group instances, these transformed representations can improve the performance of supervised learning models by removing noise and focusing on the most relevant features.
Feature Engineering:
Clusters or latent features identified through unsupervised methods can become new features in a supervised model, potentially enhancing predictive accuracy.
Beyond the Basics
Advanced unsupervised methods include:Topic Modeling:
Techniques like Latent Dirichlet Allocation (LDA) identify abstract “topics” in collections of documents.
Anomaly Detection:
Methods like Isolation Forest or Autoencoders can find unusual data points that differ significantly from the norm, useful in fraud detection or quality control.
Generative Models:
Algorithms like Generative Adversarial Networks (GANs) or Variational Autoencoders (VAEs) learn the data’s underlying distribution and can generate new, similar instances, aiding in simulation, data augmentation, or creative applications.
Why Study Unsupervised Learning
Discovering Hidden Patterns Without Labels
Learning Algorithms That Reveal Structure and Insights
Developing Skills in Exploratory Data Analysis
Understanding the Challenges and Interpretability of Results
Preparing for Advanced Research and Data-Driven Careers
Unsupervised Learning – Frequently Asked Questions
These FAQs introduce core ideas, algorithms, and study tips for unsupervised learning to support your preparation for AI and machine learning courses.
What is unsupervised learning, and how is it different from supervised learning?
Unsupervised learning is a type of machine learning where algorithms are trained on data without labeled outputs, and the goal is to discover hidden patterns, groupings, or structures in the data. In contrast, supervised learning uses labeled examples with known targets to learn a direct mapping from inputs to outputs. Unsupervised learning is especially useful when labels are expensive or unavailable, and when we want to explore data, detect clusters, or reduce dimensionality before building predictive models.
What are the main types of unsupervised learning algorithms?
The main types of unsupervised learning algorithms include clustering methods, dimensionality reduction, association rule learning, and density estimation. Clustering methods such as k-means and hierarchical clustering group similar data points together, dimensionality reduction techniques like principal component analysis (PCA) compress data into fewer informative features, and association rule learning uncovers relationships between variables in large datasets. These algorithms help simplify complex data and reveal structures that are not obvious from raw observations alone.
What are some real-world applications of unsupervised learning?
Unsupervised learning is widely used in customer segmentation, anomaly detection, recommendation systems, and exploratory data analysis. For example, businesses use clustering to group customers with similar behaviors for targeted marketing, and anomaly detection algorithms to flag unusual transactions that may indicate fraud. In recommendation systems and content platforms, unsupervised techniques help discover latent interests and organize large collections of items, making it easier to personalize user experiences.
What is clustering in unsupervised learning, and how do methods like k-means work?
Clustering is an unsupervised learning task where the goal is to group data points so that items in the same cluster are more similar to each other than to those in other clusters. In k-means clustering, we choose a number of clusters, k, and the algorithm iteratively assigns data points to the nearest cluster center and then updates the centers as the mean of their assigned points. This process continues until the assignments stabilize, producing compact clusters that summarize the main groupings in the data.
What is dimensionality reduction, and why is it important in unsupervised learning?
Dimensionality reduction refers to techniques that transform high-dimensional data into a lower-dimensional representation while preserving as much relevant information as possible. Methods such as principal component analysis (PCA), t-SNE, and autoencoders compress data into a smaller set of features that capture key patterns and variability. This is important because it helps reduce noise, visualise complex datasets, speed up learning algorithms, and mitigate the curse of dimensionality, which can make high-dimensional data difficult to analyze directly.
How can we evaluate the quality of an unsupervised learning model without labels?
Evaluating unsupervised learning is challenging because there are no ground-truth labels, but several internal and indirect measures are commonly used. For clustering, metrics such as the silhouette score, within-cluster sum of squares, and cluster separation give insight into how compact and well-separated the groups are. For dimensionality reduction and representation learning, reconstruction error, visualization quality, and downstream performance on supervised tasks can be used to judge whether the learned structure is meaningful and useful.
How does unsupervised learning support other AI and machine learning pipelines?
Unsupervised learning often acts as a foundational step in larger AI pipelines by discovering structure that can be reused in supervised tasks. It can be used for feature extraction, where patterns learned from unlabeled data provide richer inputs for later classification or regression models, and for pretraining deep networks before fine-tuning on labeled data. By organizing data, reducing dimensionality, and revealing key groups, unsupervised learning makes downstream models more efficient, robust, and informative.
What background knowledge is helpful for studying unsupervised learning at university level?
To study unsupervised learning at university level, it is helpful to have a grounding in linear algebra, probability, basic statistics, and introductory machine learning concepts. Familiarity with vector spaces, matrices, eigenvalues, and distributions makes it easier to understand algorithms like PCA and clustering. Programming experience, especially in Python and libraries such as NumPy, scikit-learn, or PyTorch, is also important for implementing algorithms and experimenting with real datasets.
Unsupervised Learning: Conclusion
Unsupervised learning turns raw, unlabeled data into structure you can use—discovering groups, compressing signals, and surfacing rare behavior without predefined labels. When paired with leakage-safe pipelines, dimensionality reduction, and (self-)supervised pretraining, it becomes a repeatable engine for discovery → hypotheses → actions.
- Understand & segment: build cluster profiles and exemplars to inform product, research, or policy decisions.
- Compress & transfer: use embeddings (PCA/UMAP/SSL) to denoise features and boost downstream models in supervised learning.
- Monitor & adapt: track drift (mix, centroids, silhouette) and retrain on schedule to keep insights trustworthy.
- Bridge to action: convert patterns into experiments, alerts, and lightweight rules; feed results back to improve the pipeline.
As data grows, unsupervised methods scale insight across domains—from computer vision and NLP to IoT and operations—complementing supervised and reinforcement approaches to build robust, adaptive AI systems.
Unsupervised Learning – Review Questions and Answers:
1. What is unsupervised learning and how does it differ from supervised learning?
Answer: Unsupervised learning is a machine learning approach where models analyze unlabeled data to discover inherent patterns, structures, or relationships without explicit guidance. Unlike supervised learning, which relies on labeled datasets to train models for specific outputs, unsupervised learning algorithms work independently to find clusters, associations, or anomalies within the data. This method is particularly useful for exploratory data analysis and feature extraction. It provides valuable insights by revealing the natural organization of data without the need for predefined classes or targets.
2. What are the primary techniques used in unsupervised learning?
Answer: The primary techniques in unsupervised learning include clustering, dimensionality reduction, and anomaly detection. Clustering algorithms such as k-means and hierarchical clustering group data points based on similarity, while dimensionality reduction techniques like Principal Component Analysis (PCA) help simplify high-dimensional data. Anomaly detection identifies unusual patterns that do not conform to expected behavior. These techniques collectively enable a comprehensive analysis of complex datasets by uncovering hidden structures and relationships.
3. How does clustering work in the context of unsupervised learning?
Answer: Clustering in unsupervised learning involves grouping data points so that those within the same cluster are more similar to each other than to those in other clusters. This process begins by selecting initial centroids and iteratively refining cluster assignments based on distance metrics. Clustering algorithms, such as k-means, then minimize the sum of squared distances between data points and their corresponding cluster centroids. Through this iterative process, the algorithm uncovers the natural groupings in the data, aiding in tasks like customer segmentation and image segmentation.
4. What is dimensionality reduction and why is it important in unsupervised learning?
Answer: Dimensionality reduction is the process of reducing the number of random variables under consideration by obtaining a set of principal variables. Techniques such as Principal Component Analysis (PCA) and t-Distributed Stochastic Neighbor Embedding (t-SNE) are commonly used for this purpose. Reducing dimensionality is important because it simplifies data visualization and speeds up the processing time while retaining the essential structure and variability of the dataset. This method also helps mitigate the curse of dimensionality, making it easier to analyze and interpret large, complex datasets.
5. How do unsupervised learning algorithms discover hidden patterns in data?
Answer: Unsupervised learning algorithms discover hidden patterns by analyzing the inherent structure of the data without relying on external labels. They use statistical techniques and distance metrics to identify clusters, correlations, and anomalies. For instance, clustering algorithms group similar data points, while dimensionality reduction methods extract the most significant features. By processing data in this exploratory manner, these algorithms reveal underlying patterns that might be overlooked using traditional analysis methods, thus offering deeper insights into the data’s structure.
6. What role does anomaly detection play in unsupervised learning?
Answer: Anomaly detection plays a crucial role in unsupervised learning by identifying data points that deviate significantly from the norm. This technique is used to detect unusual patterns, errors, or fraudulent activities in datasets without prior knowledge of what constitutes an anomaly. The process involves establishing a baseline of normal behavior and then measuring deviations from this norm using statistical or machine learning methods. Anomaly detection is essential in fields such as cybersecurity, finance, and healthcare, where early identification of outliers can prevent costly or dangerous outcomes.
7. How can unsupervised learning be used for data exploration and visualization?
Answer: Unsupervised learning facilitates data exploration and visualization by reducing complex, high-dimensional data into simpler forms that are easier to interpret. Techniques like PCA compress the data into principal components, which can then be visualized in two or three dimensions. Clustering methods help reveal the natural groupings within the data, making it possible to identify trends and patterns visually. This approach not only aids in understanding the data better but also guides further analysis and decision-making by highlighting significant relationships and structures.
8. What are some common challenges associated with unsupervised learning?
Answer: Unsupervised learning presents several challenges, including the difficulty of validating the discovered patterns without ground truth labels. Determining the optimal number of clusters or the appropriate dimensionality reduction technique can be subjective and requires careful experimentation. Noise and outliers in the data may also lead to misleading patterns, complicating the analysis. Additionally, the interpretability of the results can be challenging, as the algorithms may uncover complex structures that require domain expertise to understand fully.
9. How are evaluation metrics for unsupervised learning different from those in supervised learning?
Answer: Evaluation metrics in unsupervised learning differ significantly from those in supervised learning because there are no predefined labels to compare against. Instead of accuracy or precision, unsupervised learning relies on metrics such as silhouette score, Davies-Bouldin index, and within-cluster sum of squares (WCSS) to assess the quality of clustering and dimensionality reduction. These metrics measure the cohesion and separation of clusters or the amount of variance retained in reduced dimensions. This approach provides a quantitative way to evaluate how well the algorithm has uncovered the data’s underlying structure.
10. What are some real-world applications of unsupervised learning?
Answer: Unsupervised learning is applied in various real-world scenarios, such as market segmentation, image and speech recognition, and network anomaly detection. In market segmentation, clustering algorithms group customers based on purchasing behavior, enabling personalized marketing strategies. Dimensionality reduction techniques are used in image and speech recognition to extract meaningful features from complex data. Additionally, unsupervised learning is vital in cybersecurity for identifying unusual patterns that may indicate network intrusions or fraud, demonstrating its broad applicability across industries.
Unsupervised Learning – Thought-Provoking Questions and Answers
1. How can unsupervised learning techniques be integrated with supervised methods to enhance overall AI model performance?
Answer: Integrating unsupervised learning with supervised methods can lead to the development of hybrid models that leverage the strengths of both approaches. Unsupervised techniques, such as clustering or dimensionality reduction, can be used to preprocess and uncover hidden structures within the data before feeding it into supervised algorithms. This process can result in more robust feature extraction and improved model performance by reducing noise and redundancy. Such integration allows for the creation of more accurate and efficient models that better generalize to unseen data.
This combination not only speeds up the training process but also enhances the interpretability of the model by providing clearer insights into the underlying data patterns. By using unsupervised learning to inform feature engineering, the subsequent supervised model can focus on learning the most relevant relationships, ultimately driving better decision-making in complex applications.
2. What potential benefits and drawbacks might arise from relying solely on unsupervised learning for data analysis in complex systems?
Answer: Relying solely on unsupervised learning can offer significant benefits, such as the ability to automatically discover hidden patterns and structures in large, unlabeled datasets. This capability is particularly valuable in exploratory data analysis, where it can reveal insights that would otherwise remain hidden. The approach is also cost-effective since it does not require extensive labeled data, making it ideal for applications where labeling is impractical. However, a major drawback is the difficulty in validating and interpreting the results, as there is no ground truth to benchmark against.
Moreover, unsupervised learning algorithms can be sensitive to noise and outliers, potentially leading to misleading conclusions if not handled properly. The lack of clear evaluation metrics compared to supervised methods may also result in challenges when trying to measure model performance objectively. Thus, while unsupervised learning is powerful for uncovering structure, it is essential to combine it with expert knowledge and additional validation methods to ensure reliable insights.
3. In what ways can unsupervised learning contribute to uncovering biases hidden within large datasets?
Answer: Unsupervised learning can help uncover biases by analyzing data without predefined labels, thereby exposing inherent groupings and correlations that might be overlooked. Techniques such as clustering can reveal segments within the data that share similar characteristics, highlighting disparities or skewed distributions that indicate bias. Dimensionality reduction methods can further illuminate hidden factors influencing the data, allowing analysts to identify variables that may contribute to unfair outcomes. This process is crucial for ensuring that subsequent models built on the data are more balanced and equitable.
By identifying these biases early in the data exploration phase, organizations can take corrective measures, such as re-sampling or adjusting features, to mitigate unfairness in AI applications. The insights gained from unsupervised learning thus serve as a foundation for developing more transparent and accountable data-driven solutions, fostering trust and fairness in automated decision-making processes.
4. How might advancements in computational power influence the scalability of unsupervised learning algorithms in big data environments?
Answer: Advancements in computational power, such as the proliferation of GPUs, TPUs, and distributed computing frameworks, significantly enhance the scalability of unsupervised learning algorithms. Increased computational resources enable these algorithms to process larger datasets and more complex models in a fraction of the time required by traditional computing methods. This scalability is crucial in big data environments, where the volume and velocity of data can overwhelm conventional processing techniques. Improved hardware not only accelerates computations but also allows for the development of more sophisticated algorithms that can handle high-dimensional data more effectively.
Moreover, these technological advancements facilitate real-time data analysis, making it possible to apply unsupervised learning methods to streaming data and dynamic systems. As a result, organizations can gain timely insights and adapt their strategies quickly, driving innovation and competitive advantage in rapidly evolving markets.
5. Can unsupervised learning methods evolve to handle real-time data streams, and what are the challenges associated with that evolution?
Answer: Unsupervised learning methods can indeed evolve to handle real-time data streams through the development of online and incremental learning algorithms. These approaches update the model continuously as new data arrives, allowing for dynamic adaptation without the need to retrain from scratch. This evolution is critical in applications such as fraud detection, network monitoring, and social media analysis, where data is generated continuously and decisions must be made rapidly. The capability to process streaming data in real time ensures that models remain relevant and responsive to changing patterns and anomalies.
However, real-time unsupervised learning presents several challenges, including managing the trade-off between speed and accuracy, dealing with concept drift, and ensuring the stability of the learning process in the presence of noisy data. Efficiently updating models without compromising performance requires careful algorithm design and robust data management strategies. Addressing these challenges is essential for the successful deployment of real-time unsupervised learning systems in high-stakes environments.
6. How does the choice of distance metric impact the performance and outcomes of clustering algorithms in unsupervised learning?
Answer: The choice of distance metric is fundamental to the performance of clustering algorithms, as it directly affects how similarity between data points is quantified. Metrics such as Euclidean, Manhattan, or cosine similarity can yield markedly different clustering results depending on the structure and distribution of the data. For instance, Euclidean distance may be suitable for well-separated clusters in a continuous space, while cosine similarity might be more appropriate for high-dimensional data where orientation is more important than magnitude. The selected metric influences not only the formation of clusters but also the evaluation of cluster quality through indices like the silhouette score.
Selecting the optimal distance metric often requires domain knowledge and experimentation to understand the underlying data characteristics. A poor choice can lead to misleading groupings or the failure to detect meaningful patterns, ultimately degrading the model’s utility. Therefore, it is imperative to carefully consider and test different metrics to ensure that the clustering outcomes align with the real-world phenomena being modeled.
7. What role can unsupervised learning play in enhancing data privacy and security in the era of big data?
Answer: Unsupervised learning can enhance data privacy and security by identifying anomalous patterns that may indicate data breaches or unauthorized access. Techniques such as clustering and outlier detection enable organizations to monitor large datasets for unusual behavior without relying on pre-labeled incidents. This proactive approach allows for the early detection of security threats and the implementation of timely countermeasures. Additionally, unsupervised methods can help in segmenting data in ways that minimize the exposure of sensitive information while still extracting valuable insights.
By automating the process of anomaly detection, unsupervised learning reduces the reliance on manual monitoring and improves the overall efficiency of security systems. It also supports the development of privacy-preserving algorithms that can operate on encrypted or anonymized data, thereby maintaining compliance with data protection regulations. This dual focus on anomaly detection and data segmentation makes unsupervised learning a powerful tool for safeguarding information in increasingly complex digital environments.
8. How might unsupervised learning algorithms adapt to evolving data patterns over time without explicit re-training?
Answer: Unsupervised learning algorithms can adapt to evolving data patterns through online learning and incremental update strategies. These approaches allow models to continuously integrate new data and adjust their internal parameters dynamically, ensuring that the learned representations remain relevant as the data distribution changes. Incremental clustering algorithms, for example, can update cluster centroids in real time, while streaming PCA methods can adjust the principal components as new information becomes available. This adaptability is essential in environments where data evolves rapidly, such as social media analytics or sensor networks.
By incorporating mechanisms for continual learning, unsupervised algorithms minimize the need for periodic complete re-training, thereby reducing computational overhead and downtime. This capability not only maintains model performance in the face of concept drift but also enables timely insights into emerging trends. The ability to self-adapt is a key factor in the long-term effectiveness of unsupervised learning systems in dynamic and complex data ecosystems.
9. What are the implications of unsupervised learning for personalized recommendation systems in dynamic online environments?
Answer: Unsupervised learning has significant implications for personalized recommendation systems, as it enables the discovery of user behavior patterns and preferences without relying on explicit ratings or feedback. By clustering users based on browsing history, purchase behavior, or content interaction, recommendation systems can identify similar user groups and tailor suggestions accordingly. This approach allows for the development of dynamic models that adapt to evolving user interests and market trends, resulting in more relevant and timely recommendations. The flexibility of unsupervised methods also facilitates the integration of diverse data sources, enriching the personalization process.
In dynamic online environments, where user behavior can change rapidly, unsupervised learning helps maintain the freshness and accuracy of recommendations. It empowers systems to detect emerging trends and niche interests, thereby enhancing user engagement and satisfaction. Ultimately, the incorporation of unsupervised techniques into recommendation engines drives innovation in personalized content delivery and customer experience.
10. How can visualizations derived from unsupervised learning insights improve decision-making in business intelligence?
Answer: Visualizations generated from unsupervised learning insights, such as cluster plots and heat maps, can significantly enhance decision-making by revealing underlying data patterns and trends. These visual tools help stakeholders quickly grasp complex relationships and identify critical areas that require attention. For example, a well-designed cluster visualization can highlight customer segments with distinct purchasing behaviors, enabling targeted marketing strategies. By translating high-dimensional data into intuitive graphical representations, unsupervised learning makes data-driven insights more accessible and actionable.
Such visualizations not only aid in strategic planning but also foster a deeper understanding of the business landscape. They serve as a bridge between complex data analytics and practical decision-making, empowering leaders to make informed choices that drive growth and efficiency. In this way, the integration of unsupervised learning with data visualization techniques transforms raw data into strategic business intelligence.
11. In what ways can unsupervised learning be applied to natural language processing to uncover semantic relationships in text data?
Answer: Unsupervised learning can be applied to natural language processing (NLP) through techniques such as topic modeling, word embeddings, and clustering of text documents. These methods uncover semantic relationships by analyzing patterns in word co-occurrence and document similarity, allowing for the discovery of latent topics within large corpora. For instance, algorithms like Latent Dirichlet Allocation (LDA) can identify topics that represent recurring themes across texts without prior labeling. Similarly, unsupervised word embedding techniques such as Word2Vec capture contextual relationships between words, enabling a deeper understanding of language semantics.
By extracting these hidden structures, unsupervised NLP methods facilitate tasks such as sentiment analysis, information retrieval, and automated summarization. The insights generated can help organizations understand customer feedback, monitor trends, and improve content recommendations. This unsupervised approach to language understanding is particularly valuable in scenarios where labeled data is scarce, making it a powerful tool for modern text analytics.
12. How might the integration of unsupervised learning with emerging fields such as quantum computing revolutionize data analysis?
Answer: The integration of unsupervised learning with emerging technologies like quantum computing holds the promise of revolutionizing data analysis by dramatically accelerating the processing of complex, high-dimensional datasets. Quantum computing can perform certain computations exponentially faster than classical computers, which may enable unsupervised algorithms to tackle previously intractable problems in clustering and dimensionality reduction. This synergy could lead to breakthroughs in pattern recognition and anomaly detection, unlocking new levels of efficiency and accuracy in data-driven decision-making.
Moreover, quantum-enhanced unsupervised learning could transform fields such as genomics, finance, and cybersecurity by providing unprecedented insights into vast and complex datasets. The ability to process and analyze data in real time using quantum algorithms would pave the way for more adaptive and responsive systems. As quantum technologies mature, their integration with unsupervised learning is expected to redefine the boundaries of what is achievable in data analysis and artificial intelligence.
Unsupervised Learning -Numerical Problems and Solutions
1. Euclidean Distance Calculation in Clustering
Solution:
Step 1: Given two data points A = (2, -1, 3) and B = (5, 1, 0), compute the difference for each coordinate: (5–2, 1–(–1), 0–3) = (3, 2, -3).
Step 2: Square each difference: 3² = 9, 2² = 4, (–3)² = 9.
Step 3: Sum the squared differences and take the square root: √(9 + 4 + 9) = √22 ≈ 4.69.
2. Sum of Squared Errors (SSE) for K-Means Clustering
Solution:
Step 1: For data points (2,3), (3,4), (4,5) and centroid (3,4), calculate the squared Euclidean distance for each point.
Step 2: Compute distances: For (2,3): (2–3)²+(3–4)² = 1+1 = 2; for (3,4): (0)²+(0)² = 0; for (4,5): (4–3)²+(5–4)² = 1+1 = 2.
Step 3: Sum these squared errors: SSE = 2 + 0 + 2 = 4.
3. Silhouette Score Calculation
Solution:
Step 1: Given an average intra-cluster distance (a) of 2.5 and a nearest-cluster distance (b) of 4.0, compute the difference: b – a = 4.0 – 2.5 = 1.5.
Step 2: Identify the maximum of a and b, which is 4.0.
Step 3: Calculate the silhouette score: 1.5 ÷ 4.0 = 0.375.
4. Variance Explained by Principal Component Analysis (PCA)
Solution:
Step 1: Given eigenvalues λ₁ = 5, λ₂ = 3, and λ₃ = 2, compute the total variance: 5 + 3 + 2 = 10.
Step 2: Determine the variance explained by the first principal component: 5 ÷ 10 = 0.5.
Step 3: Express the result as a percentage: 0.5 × 100 = 50%.
5. Cosine Similarity Calculation Between Two Vectors
Solution:
Step 1: Given vectors A = (1,2,3) and B = (4,5,6), compute the dot product: (1×4) + (2×5) + (3×6) = 4 + 10 + 18 = 32.
Step 2: Calculate the magnitude of each vector: ||A|| = √(1²+2²+3²) = √14 and ||B|| = √(4²+5²+6²) = √77.
Step 3: Compute the cosine similarity: 32 ÷ (√14 × √77) ≈ 32 ÷ (√1078) ≈ 32 ÷ 32.84 ≈ 0.974.
6. Dunn Index Calculation for Cluster Validation
Solution:
Step 1: Given the minimum inter-cluster distance is 3.5 and the maximum intra-cluster distance is 1.2, set up the ratio: (3.5) ÷ (1.2).
Step 2: Perform the division: 3.5 ÷ 1.2 ≈ 2.9167.
Step 3: Conclude that the Dunn index is approximately 2.92, indicating well-separated clusters.
7. Adjusted Rand Index (ARI) Calculation
Solution:
Step 1: For a contingency matrix [[2, 1], [1, 2]], compute the sum over each cell’s combination: C(2,2) + C(1,2) + C(1,2) + C(2,2) = 1 + 0 + 0 + 1 = 2.
Step 2: Compute the sum of combinations for row sums: Row1 sum = 3 → C(3,2) = 3; Row2 sum = 3 → C(3,2) = 3; total = 6; and similarly for column sums (total = 6).
Step 3: Calculate total pairs: C(6,2) = 15. Then, ARI = (2 – (6×6)/15) ÷ (0.5×(6+6) – (6×6)/15) = (2 – 2.4) ÷ (6 – 2.4) = (–0.4) ÷ 3.6 ≈ –0.1111.
8. Davies-Bouldin Index Calculation for Two Clusters
Solution:
Step 1: Given average intra-cluster distances of 1.5 and 1.2 for clusters 1 and 2, compute their sum: 1.5 + 1.2 = 2.7.
Step 2: Given the inter-cluster distance is 4.0, set up the ratio: 2.7 ÷ 4.0.
Step 3: Calculate the Davies-Bouldin index: 2.7 ÷ 4.0 = 0.675.
9. Principal Component Projection of a Data Point
Solution:
Step 1: Given a data point (2, 3) and a normalized first principal component vector v = (0.6, 0.8), compute the dot product: (2×0.6) + (3×0.8).
Step 2: Multiply: 2×0.6 = 1.2 and 3×0.8 = 2.4.
Step 3: Sum the products to obtain the projection: 1.2 + 2.4 = 3.6.
10. Total Within-Cluster Sum of Squares (WCSS) Calculation
Solution:
Step 1: For a cluster with centroid (3,3) and data points (2,2), (3,4), (4,3), compute the squared distance for each point from the centroid.
Step 2: For (2,2): (1² + 1²) = 2; for (3,4): (0² + 1²) = 1; for (4,3): (1² + 0²) = 1.
Step 3: Sum the squared distances: 2 + 1 + 1 = 4.
11. Eigenvalue Calculation for a 2×2 Covariance Matrix
Solution:
Step 1: Given the covariance matrix [[4, 2], [2, 3]], set up the characteristic equation by subtracting λ from the diagonal: det([[4–λ, 2], [2, 3–λ]]) = (4–λ)(3–λ) – (2×2).
Step 2: Expand the determinant: (4–λ)(3–λ) = 12 – 4λ – 3λ + λ² = λ² – 7λ + 12; subtract 4 to get λ² – 7λ + 8 = 0.
Step 3: Solve the quadratic equation λ² – 7λ + 8 = 0 using the quadratic formula: λ = [7 ± √(49 – 32)] ÷ 2 = [7 ± √17] ÷ 2.
12. Dimensionality Reduction Percentage Using PCA
Solution:
Step 1: Given the original dimensionality of 50 and a reduced dimensionality of 10, calculate the reduction in dimensions: 50 – 10 = 40.
Step 2: Determine the reduction ratio: 40 ÷ 50 = 0.8.
Step 3: Express the result as a percentage: 0.8 × 100 = 80% reduction in dimensionality.
